OCR Specification focus:
‘Outline features used to classify Prokaryotae, Protoctista, Fungi, Plantae and Animalia; discuss molecular evidence leading to the three-domain model.’
Classification in biology organises life’s immense diversity into meaningful groups. Understanding how organisms are classified reveals evolutionary relationships and how modern evidence reshapes traditional biological systems.
The Five Kingdoms of Life
Biological classification historically divided all living organisms into five kingdoms, based primarily on cell structure, mode of nutrition, and organisation. These kingdoms—Prokaryotae, Protoctista, Fungi, Plantae, and Animalia—reflect key differences in cellular complexity and life processes.
Limitations of the Five-Kingdom System
While the five-kingdom model simplified classification, it has several limitations:
Overgeneralisation: The group Protoctista includes organisms that are only loosely related.
Prokaryote diversity: Molecular studies revealed significant genetic and biochemical differences between types of prokaryotes, such as bacteria and archaea.
Evolutionary ambiguity: The model does not accurately reflect evolutionary relationships or genetic ancestry.
These shortcomings led scientists to seek a more precise system based on molecular data rather than observable characteristics alone.
Molecular Evidence and the Three-Domain Model
With advances in molecular biology and genetic sequencing, scientists began comparing ribosomal RNA (rRNA) sequences across organisms. rRNA evolves slowly, making it ideal for tracing long-term evolutionary divergence.
Discovery of Archaea
Comparisons revealed that some prokaryotes, previously grouped together, were genetically distinct. These new organisms were named Archaea.
Archaea: A group of single-celled microorganisms genetically distinct from bacteria and often living in extreme environments.
Archaea differ from Bacteria in key ways:
Cell membrane lipids are chemically distinct.
Cell walls lack peptidoglycan.
Genes and enzymes resemble those of eukaryotes more closely than bacteria.
Many are extremophiles, thriving in high-salt, high-temperature, or acidic environments.
This discovery transformed understanding of evolutionary relationships, prompting a major reorganisation of classification.
The Three Domains
Biologist Carl Woese proposed the three-domain model in 1977, refining it in 1990 using rRNA sequences to separate Bacteria, Archaea and Eukarya.
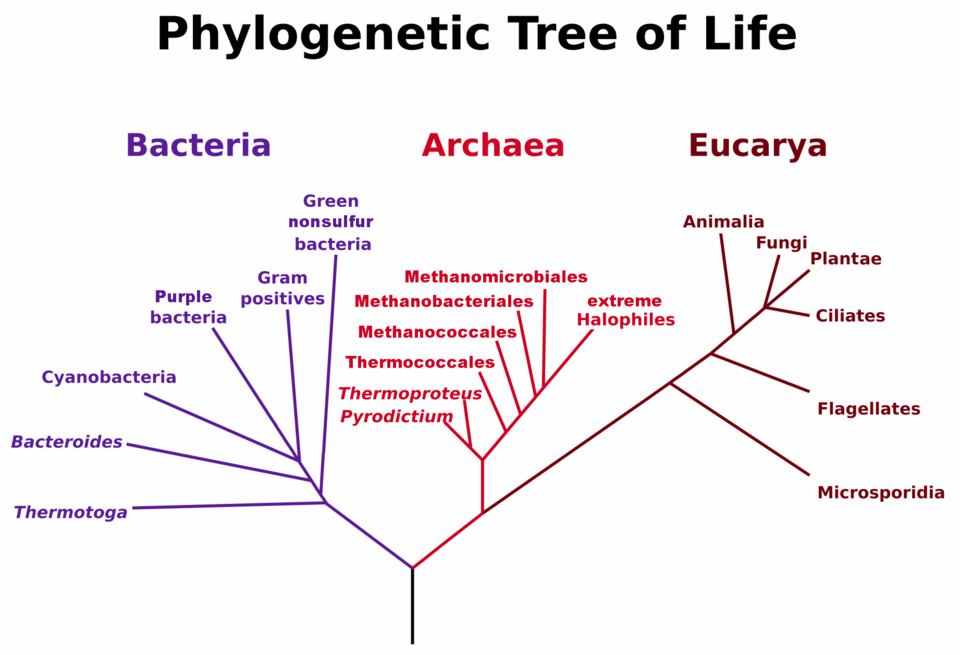
Universal phylogenetic tree (after Woese et al., 1990) based on rRNA sequence comparisons. It depicts the three primary domains—Bacteria, Archaea, and Eukarya—showing their deep evolutionary divergence. Some fine branch details exceed OCR depth, but the domain-level division is the essential concept. Source.
The three domains are Bacteria, Archaea and Eukarya, which represent the deepest split supported by molecular data.
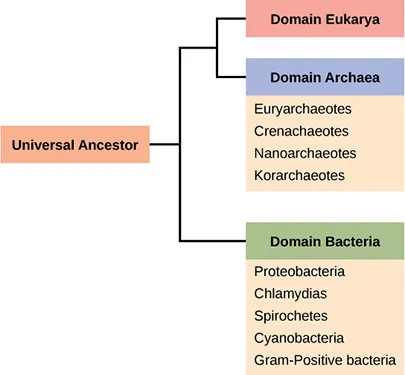
OpenStax Figure 22.11 presents a simplified schematic of the three domains of life—Bacteria, Archaea, and Eukarya—illustrating how molecular data reshaped classification. The surrounding page adds prokaryote structure details not required by the OCR syllabus, but the figure itself directly reinforces domain-level understanding. Source.
These domains represent the highest taxonomic rank, above kingdom, and are based primarily on molecular evidence, not morphology.
The model highlights that Archaea and Eukarya share a more recent common ancestor than either does with Bacteria, suggesting eukaryotes evolved from archaean lineages.
Modern Implications of Molecular Classification
Molecular data, including DNA sequencing and genomic analysis, now underpin modern taxonomy. This has reshaped understanding of evolutionary pathways and species relationships.
Key advances include:
Use of phylogenetic trees derived from genetic data to illustrate evolutionary descent.
Identification of cryptic species previously indistinguishable morphologically.
Reclassification of organisms into clades based on genetic similarity rather than physical traits.
Clarification of horizontal gene transfer among prokaryotes, revealing complex evolutionary histories.
Modern classification continues to evolve as technology advances, providing ever-greater precision in mapping the diversity and unity of life.
FAQ
Scientists use ribosomal RNA (rRNA) sequencing, DNA hybridisation, and whole-genome analysis to assess genetic similarity.
rRNA is particularly useful because it is present in all organisms, evolves slowly, and shows conserved regions suitable for comparison.
Other molecular tools include:
PCR (Polymerase Chain Reaction) to amplify rRNA genes.
Bioinformatics software to align sequences and measure genetic distance.
These comparisons help reveal evolutionary relationships beyond visible characteristics.
rRNA genes are universally present and perform an essential cellular function, so they are conserved across all life forms.
Their slow rate of evolution allows scientists to detect deep evolutionary relationships, while variable regions offer clues about more recent divergence.
This balance of conservation and variability makes rRNA ideal for identifying differences between Bacteria, Archaea, and Eukarya.
Archaeal membranes are unique because:
They contain ether-linked lipids, whereas bacterial and eukaryotic membranes have ester-linked lipids.
The lipid tails are often branched isoprenoid chains, increasing stability.
Archaea may form monolayers instead of bilayers, enhancing resistance to extreme heat or pH.
These adaptations explain how many Archaea survive in environments like hot springs or salt lakes.
No. Although many are extremophiles, living in hot, salty, or acidic conditions, others inhabit moderate environments such as soil, oceans, and animal digestive tracts.
Recent genomic research shows that non-extreme Archaea play key ecological roles, such as:
Methane cycling in wetlands and marine sediments.
Ammonia oxidation in nutrient cycling.
Their diversity and ubiquity were underestimated before molecular methods became available.
The three-domain model reshaped the tree of life into a structure showing two distinct prokaryotic lineages (Bacteria and Archaea) and one eukaryotic lineage (Eukarya).
This model emphasises that Eukarya and Archaea share a more recent common ancestor, challenging the earlier assumption that all prokaryotes formed a single, primitive group.
It also highlights that the evolution of complex cells likely arose from symbiotic relationships between early archaean and bacterial cells.
Practice Questions
Question 1 (2 marks)
State two structural or biochemical differences between members of the domains Bacteria and Archaea.
Mark Scheme:
Award 1 mark for each correct difference, up to 2 marks total.
Bacteria have cell walls containing peptidoglycan; Archaea lack peptidoglycan in their cell walls. (1 mark)
Membrane lipids in Archaea contain ether bonds, whereas those in Bacteria contain ester bonds. (1 mark)
Accept any other valid biochemical or structural distinction such as differences in rRNA sequences or sensitivity to antibiotics (max 2).
Question 2 (5 marks)
Describe how molecular evidence has led to the development of the three-domain classification system and explain how it differs from the five-kingdom model.
Mark Scheme:
Award marks for any five of the following points:
Comparison of rRNA or DNA sequences revealed fundamental differences between types of prokaryotes. (1 mark)
Discovery of Archaea as a distinct group, separate from Bacteria, based on molecular data. (1 mark)
Carl Woese proposed the three-domain model (Bacteria, Archaea, Eukarya) following rRNA studies. (1 mark)
The three-domain system is based on molecular and genetic evidence, not just observable characteristics. (1 mark)
The five-kingdom model grouped all prokaryotes together (in Prokaryotae/Monera) and was based mainly on morphology and metabolism. (1 mark)
In the three-domain system, Eukarya encompasses Protoctista, Fungi, Plantae, and Animalia as separate kingdoms under one domain. (1 mark, max 5 total)
Indicative content: A well-structured answer should show understanding that molecular evidence (especially rRNA sequencing) provided a more accurate reflection of evolutionary relationships, leading to the separation of Archaea from Bacteria and a higher-level classification above kingdoms.

