OCR Specification focus:
‘Outline roles of RNA polymerase, mRNA, tRNA, and rRNA in polypeptide synthesis.’
The synthesis of proteins through transcription and translation converts genetic information in DNA into functional polypeptides, involving the coordinated roles of RNA molecules and ribosomal components.
Transcription: Copying the Genetic Code
Overview of Transcription
Transcription is the process by which a gene’s DNA sequence is copied into a complementary messenger RNA (mRNA) molecule. This occurs in the nucleus of eukaryotic cells. The resulting mRNA carries the genetic instructions to the cytoplasm for translation.
RNA Polymerase: The Key Enzyme
RNA polymerase catalyzes the synthesis of mRNA from the DNA template.
RNA Polymerase: An enzyme that binds to DNA and synthesizes a strand of mRNA using complementary base pairing rules.
Stages of Transcription:
Initiation:
The enzyme RNA polymerase binds to a specific region called the promoter on the DNA.
The DNA double helix unwinds, exposing the template strand.
Elongation:
RNA polymerase moves along the DNA template strand, synthesizing an mRNA molecule by joining ribonucleotides through phosphodiester bonds.
Base pairing occurs between DNA and RNA nucleotides:
Adenine (A) pairs with Uracil (U), not Thymine.
Cytosine (C) pairs with Guanine (G).
Termination:
When the polymerase reaches a termination sequence, it detaches from the DNA, and the newly formed pre-mRNA is released.
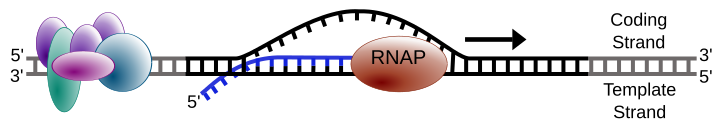
A simplified diagram of transcription elongation, with RNA polymerase tracking the DNA template strand to produce a complementary RNA transcript. Labels distinguish the coding vs template strands and the nascent RNA’s 5′→3′ growth. The minimal layout avoids extraneous regulatory detail, matching the syllabus's focus on roles. Source.
Post-Transcriptional Modifications
In eukaryotes, the initial pre-mRNA undergoes several modifications before leaving the nucleus:
Splicing: Removal of introns (non-coding regions) and joining of exons (coding regions).
5′ cap addition: A modified guanine nucleotide is added to the 5′ end for stability and ribosome recognition.
Poly-A tail addition: A long chain of adenine nucleotides added to the 3′ end, protecting mRNA from degradation.
After these modifications, the mature mRNA exits the nucleus via the nuclear pore and enters the cytoplasm.
The Role of Messenger RNA (mRNA)
mRNA acts as the temporary copy of a gene that directs polypeptide synthesis at ribosomes.
Messenger RNA (mRNA): A single-stranded RNA molecule that carries the genetic code from DNA to the ribosome for translation.
Each sequence of three nucleotides in mRNA is known as a codon, which corresponds to one amino acid in the protein. The sequence of codons determines the order of amino acids in the resulting polypeptide.
Translation: Converting the Code into a Polypeptide
Overview of Translation
Translation is the process that decodes the information carried by mRNA to assemble a polypeptide chain. It occurs at the ribosome in the cytoplasm.
Ribosomes and Ribosomal RNA (rRNA)
Ribosomes are the sites of protein synthesis, composed of two subunits: one large and one small. Each subunit contains rRNA and proteins.
Ribosomal RNA (rRNA): A structural and catalytic component of ribosomes that ensures correct alignment of mRNA and tRNA and catalyzes peptide bond formation.
rRNA forms the peptidyl transferase center, the site of peptide bond formation between amino acids.
The Role of Transfer RNA (tRNA)
tRNA molecules deliver specific amino acids to the ribosome based on the codons on the mRNA strand.
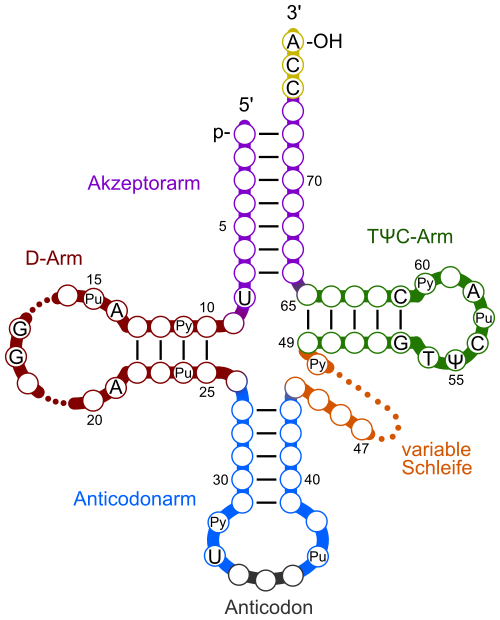
The cloverleaf model of tRNA highlights the anticodon loop that base-pairs with mRNA codons and the 3′ acceptor stem (CCA) where the amino acid attaches. Extra loops (D and TψC) and the variable loop are shown; these details exceed the syllabus but aid orientation. Labels are clear, and the SVG scales cleanly for web use. Source.
Transfer RNA (tRNA): A small RNA molecule that carries a specific amino acid and recognizes codons on mRNA through its complementary anticodon.
Each tRNA has:
An anticodon loop that pairs with a complementary codon on the mRNA.
An amino acid attachment site corresponding to that anticodon.
Charging of tRNA
Each tRNA molecule becomes aminoacylated when its specific amino acid is attached by an enzyme called aminoacyl-tRNA synthetase. This activation step requires ATP.
The Process of Translation
Translation proceeds in three key stages: initiation, elongation, and termination.
Initiation
The small ribosomal subunit binds to the 5′ end of the mRNA.
The start codon (AUG) is recognized by the initiator tRNA, carrying methionine.
The large ribosomal subunit attaches, forming a complete ribosome.
Elongation
A new tRNA molecule carrying the next amino acid binds to the A site of the ribosome, matching its anticodon to the mRNA codon.
A peptide bond forms between the amino acid at the P site and the new amino acid at the A site, catalyzed by rRNA.
The ribosome then shifts (translocates) one codon along the mRNA, moving the empty tRNA to the E site for release.
Termination
When a stop codon (UAA, UAG, or UGA) is reached, no tRNA binds.
Instead, a release factor binds to the stop codon, triggering release of the completed polypeptide chain.
The ribosome subunits dissociate, ready to begin another translation cycle.
Polypeptide Processing
Following translation, the polypeptide may undergo post-translational modifications, such as:
Folding into a specific tertiary structure is aided by chaperone proteins.
Cleavage of initial amino acids or signal peptides.
Chemical modification, such as phosphorylation or glycosylation, is used to activate or target the protein.
These modifications ensure the protein achieves its functional conformation and correct cellular destination.
Integration of Transcription and Translation
In prokaryotes, transcription and translation occur simultaneously in the cytoplasm because there is no nuclear membrane separating the processes.
In eukaryotes, transcription occurs in the nucleus, and translation in the cytoplasm — creating a spatial and temporal separation that allows RNA processing to occur.
FAQ
In prokaryotes, there is no nuclear envelope separating DNA from ribosomes. As soon as an mRNA strand begins to form, ribosomes can attach and start translation — this is known as coupled transcription and translation.
In eukaryotes, transcription occurs inside the nucleus, and mRNA must be processed (splicing, capping, tailing) before it exits to the cytoplasm for translation. The spatial separation of these processes prevents them from occurring simultaneously.
RNA polymerase identifies a specific region on the DNA called the promoter. Promoters contain conserved sequences, such as the TATA box, that signal where transcription should begin.
In eukaryotes, transcription factors bind to the promoter first, helping RNA polymerase position correctly. In prokaryotes, a sigma factor performs a similar role, ensuring initiation occurs at the correct site on the DNA template.
A codon of three bases provides enough combinations to encode all 20 amino acids using only four nucleotide bases (A, U, C, G).
Two-base codons would only yield 16 possible combinations, which is insufficient.
Four-base codons would create 256 combinations, introducing unnecessary redundancy.
Thus, the triplet code represents an optimal balance of efficiency and accuracy in translation.
Each tRNA is charged with an amino acid by a specific aminoacyl-tRNA synthetase. If an incorrect amino acid is attached, the ribosome will still add it to the growing polypeptide, as it recognizes only the anticodon–codon pairing, not the amino acid itself.
This rare error leads to a misfolded or dysfunctional protein, but proofreading by the synthetase minimizes such mistakes, maintaining high translation fidelity.
Release factors are proteins that recognize stop codons (UAA, UAG, UGA) on the mRNA during translation.
When a stop codon enters the A site of the ribosome, no corresponding tRNA binds.
Instead, a release factor binds to the stop codon.
This action triggers hydrolysis of the bond linking the polypeptide to the tRNA in the P site, freeing the finished protein.
The ribosomal subunits then dissociate, completing translation.
Practice Questions
Question 1 (2 marks)
Describe the role of RNA polymerase in transcription.
Mark scheme:
(1 mark) RNA polymerase binds to the DNA template strand at the promoter region.
(1 mark) It catalyzes the formation of phosphodiester bonds between complementary RNA nucleotides, producing a strand of mRNA.
Question 2 (5 marks)
Explain how the structures of mRNA, tRNA, and rRNA enable them to carry out their functions during translation.
Mark scheme:
(1 mark) mRNA is a single-stranded molecule carrying the sequence of codons that determines the order of amino acids in the polypeptide.
(1 mark) Its short lifespan and structure allow it to pass through the nuclear pore and attach to the ribosome in the cytoplasm.
(1 mark) tRNA has an anticodon loop that base-pairs with a complementary codon on mRNA.
(1 mark) tRNA has an amino acid attachment site (acceptor stem) for adding specific amino acids.
(1 mark) rRNA forms part of the ribosome’s catalytic site, aligning mRNA and tRNA and catalyzing peptide bond formation between amino acids.

