OCR Specification focus:
‘Outline how primary mRNA is edited, including intron removal and exon splicing, to produce mature mRNA prior to translation.’
In eukaryotic cells, gene expression requires precise modification of RNA transcripts. Post-transcriptional control through mRNA processing ensures accurate, functional messenger RNA is produced for translation, enabling protein synthesis regulation.
Post-Transcriptional Control Overview
After transcription, the initial RNA molecule formed is called primary mRNA or pre-mRNA. This molecule contains both exons (coding regions) and introns (non-coding regions). Before translation can occur, the pre-mRNA must be processed to form mature mRNA, which is then exported from the nucleus to the cytoplasm. Post-transcriptional control regulates gene expression by determining which mRNA transcripts are available for translation, influencing the quantity and diversity of proteins produced.
The Stages of mRNA Processing
1. Addition of the 5′ Cap
A 5′ cap is added to the start of the pre-mRNA shortly after transcription begins. This modification involves attachment of a methylated guanine nucleotide to the 5′ end via a triphosphate bridge.
Protects the mRNA from degradation by exonucleases.
Facilitates ribosome binding during translation initiation.
Aids in nuclear export of the mRNA molecule.
The capping process is vital to stabilise the transcript and prepare it for later stages of gene expression.
2. Addition of the Poly-A Tail
At the 3′ end of the pre-mRNA, a long chain of adenine nucleotides—known as the poly-A tail—is added by the enzyme poly-A polymerase.
Protects the mRNA from enzymatic degradation in the cytoplasm.
Assists in the termination of transcription.
Enhances translation efficiency by aiding ribosome recycling.
The length of the poly-A tail can vary between transcripts, affecting the mRNA’s stability and lifespan, thereby influencing protein synthesis levels.
3. RNA Splicing
RNA splicing is the process by which introns are removed and exons joined together to create a continuous coding sequence in the mature mRNA.
Intron: A non-coding sequence of DNA or RNA that is removed during RNA processing and does not contribute to the final protein sequence.
Exon: A coding sequence of DNA or RNA that remains in the processed mRNA and is expressed as part of the protein sequence.
Splicing is carried out by a large molecular complex called the spliceosome, composed of small nuclear RNAs (snRNAs) and proteins. The spliceosome recognises specific nucleotide sequences at the intron–exon boundaries and catalyses the two key transesterification reactions necessary for intron removal.
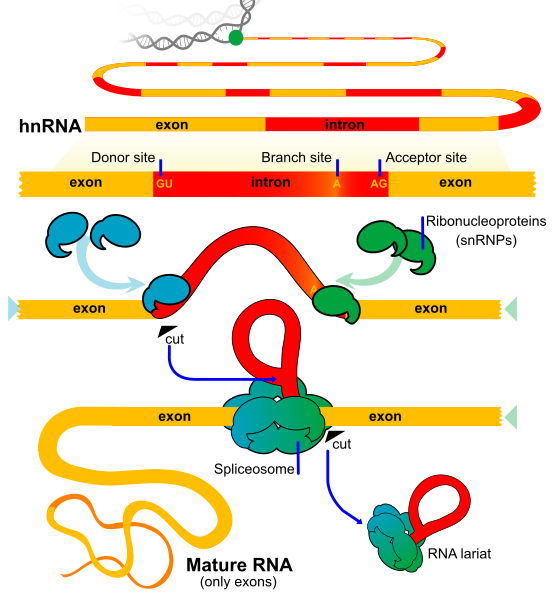
A labelled diagram of RNA splicing showing exons retained and introns removed as a lariat, mediated by the spliceosome. This visual supports recognition of conserved splice sites and the joining of exons into a continuous coding sequence. The layout is uncluttered and suitable for A-level study. Source.
The stages include:
Recognition of the 5′ and 3′ splice sites and branch point.
Lariat formation, where the intron loops back on itself.
Exon ligation, joining adjacent exons to form a continuous mRNA strand.
This process ensures that the mRNA sequence corresponds accurately to the gene’s coding regions.
4. Alternative Splicing
Alternative splicing allows a single gene to give rise to multiple protein variants, significantly increasing proteomic complexity.
Different combinations of exons may be included or excluded.
This leads to different isoforms of a protein with varied functions or properties.
Regulation occurs through splicing factors, which promote or repress specific splice sites.
For example, in human muscle tissue, alternative splicing allows the production of different forms of tropomyosin, suited to distinct cell types.
Alternative splicing: The regulated process by which different combinations of exons are joined during RNA splicing, resulting in the production of multiple mRNA variants from a single gene.
This mechanism exemplifies the flexibility of eukaryotic gene expression and its control beyond DNA transcription.
Molecular Machinery in mRNA Processing
The Spliceosome Complex
The spliceosome comprises five small nuclear ribonucleoproteins (snRNPs): U1, U2, U4, U5, and U6. These recognise conserved sequences—typically the GU sequence at the 5′ end of introns and the AG sequence at the 3′ end. Coordinated interactions between these components ensure precise and efficient splicing.
Key roles of snRNPs:
U1 snRNP binds to the 5′ splice site.
U2 snRNP binds near the branch point adenine.
U4/U6.U5 tri-snRNP complex mediates intron looping and exon joining.
Errors in splicing can result in aberrant mRNAs, potentially leading to non-functional or harmful proteins, and are associated with genetic diseases such as spinal muscular atrophy.
RNA Editing and Quality Control
After splicing, further editing may occur to modify specific nucleotides, changing the coding potential of the mRNA.
RNA editing: A molecular process in which nucleotide bases in an RNA molecule are altered, inserted, or deleted after transcription, modifying the mRNA sequence without changing the underlying DNA.
Editing enables functional diversity and regulatory control, for example by altering codons to generate different amino acids. Following processing, quality control mechanisms—such as the nuclear exosome and nonsense-mediated decay (NMD)—identify and degrade defective mRNAs, preventing production of abnormal proteins.
Export of Mature mRNA
Once processing is complete, the mature mRNA—now capped, polyadenylated, and spliced—is exported from the nucleus through nuclear pores to the cytoplasm. Specific export proteins recognise the cap and tail, ensuring only correctly processed mRNAs leave the nucleus. Once in the cytoplasm, ribosomes bind to the 5′ cap to initiate translation, converting the mRNA sequence into a polypeptide chain.

A schematic of a mature eukaryotic mRNA, highlighting the 5′ cap, coding region, 3′ poly-A tail, and UTRs. It succinctly visualises the product of post-transcriptional editing prior to translation. Note: UTR labelling provides useful context beyond the strict OCR wording but does not add unnecessary complexity. Source.
Significance of Post-Transcriptional Control
Post-transcriptional control enables cells to:
Regulate gene expression rapidly without altering transcription rates.
Produce multiple proteins from a single gene through alternative splicing.
Maintain quality assurance in mRNA molecules.
Control protein synthesis in response to developmental or environmental signals.
This fine-tuned control over mRNA maturation is essential for cell differentiation, organismal development, and adaptive responses, ensuring that the proteome accurately reflects cellular needs and environmental conditions.
FAQ
Splicing occurs at specific nucleotide sequences recognised by the spliceosome. These include:
A GU sequence at the 5′ splice site (beginning of the intron).
An AG sequence at the 3′ splice site (end of the intron).
A branch point adenine (A) located within the intron, used to form the lariat structure.
The precise recognition of these sites ensures accurate removal of introns and correct assembly of exons into mature mRNA.
Alternative splicing allows different combinations of exons to be retained or excluded from the final mRNA.
This means a single gene can generate multiple mRNA transcripts, each coding for a distinct protein isoform with variations in amino acid sequence or function.
For example, in humans, over 90% of genes undergo alternative splicing, explaining how a limited number of genes can produce a vastly diverse proteome.
The 5′ capping process uses guanylyl transferase, which attaches a modified guanine nucleotide, followed by methyltransferase, which adds a methyl group.
For the poly-A tail, an enzyme called poly-A polymerase adds adenine nucleotides after cleavage at the 3′ end of the pre-mRNA.
These modifications protect mRNA from degradation and assist in translation initiation and nuclear export.
mRNA quality control ensures only correctly processed transcripts are translated.
Nonsense-mediated decay (NMD) eliminates mRNAs containing premature stop codons.
The nuclear exosome degrades defective or unspliced RNA.
Export control proteins block faulty mRNAs from leaving the nucleus.
These mechanisms prevent synthesis of abnormal or harmful proteins, maintaining cellular integrity.
Faulty splicing can lead to inclusion of introns or exclusion of essential exons, altering the protein sequence.
For example:
Spinal muscular atrophy (SMA) results from incorrect splicing of the SMN1 gene, reducing functional protein levels.
Beta-thalassaemia can occur when splicing errors disrupt haemoglobin gene expression.
Such mutations highlight the importance of precise post-transcriptional control in maintaining normal gene function.
Practice Questions
Question 1 (2 marks)
Describe two ways in which eukaryotic pre-mRNA is modified before translation.
Mark scheme:
Addition of a 5′ cap to the beginning of the mRNA (1 mark)
Addition of a poly-A tail to the 3′ end (1 mark)
Splicing to remove introns and join exons (1 mark, maximum 2 marks total)
Question 2 (5 marks)
Explain how RNA splicing and alternative splicing contribute to gene expression and protein diversity in eukaryotic cells.
Mark scheme:
Splicing removes non-coding introns from pre-mRNA (1 mark)
Exons are joined together to form the mature mRNA (1 mark)
This ensures only coding sequences are translated into protein (1 mark)
Alternative splicing allows different combinations of exons to be included or excluded (1 mark)
This produces different mRNA transcripts from the same gene, leading to different protein isoforms with varied functions (1 mark)

