OCR Specification focus:
‘Explain the polymerase chain reaction and its application in amplifying DNA for analysis and diagnostics.’
The Polymerase Chain Reaction (PCR): Principles and Applications
PCR is a molecular technique used to amplify specific DNA sequences rapidly and accurately, enabling millions of copies to be produced from minute samples for analysis and diagnostics.
Principle of PCR
Overview of DNA Amplification
PCR (Polymerase Chain Reaction) is an in vitro method for amplifying DNA. It mimics natural DNA replication but occurs in a controlled laboratory setting using temperature cycles and enzymes. The process relies on DNA polymerase, primers, and nucleotides to replicate a target sequence exponentially.
DNA Polymerase: An enzyme that catalyses the synthesis of new DNA strands from deoxyribonucleotide triphosphates using a DNA template.
PCR was developed by Kary Mullis in 1983, revolutionising molecular biology and genetics by allowing DNA amplification without the need for living cells. It underpins techniques used in genetic testing, forensics, and medical diagnostics.
Key Components of PCR
Essential Reagents
PCR requires several core components:
Template DNA – the sample containing the target sequence to be amplified.
Primers – short, single-stranded DNA sequences (around 20 bases) that are complementary to regions flanking the target sequence.
DNA polymerase – a thermostable enzyme such as Taq polymerase (from Thermus aquaticus) capable of withstanding high temperatures.
Deoxyribonucleotide triphosphates (dNTPs) – the building blocks (A, T, C, G) for new DNA strand synthesis.
Buffer solution and magnesium ions – to maintain pH and provide cofactors for enzyme activity.
Taq Polymerase: A heat-stable DNA polymerase derived from Thermus aquaticus, used in PCR to synthesise DNA at high temperatures without denaturation.
The PCR Cycle
Step-by-Step Process
PCR operates in repeated temperature-controlled steps, usually within a thermocycler. Each cycle doubles the DNA quantity, leading to exponential amplification.
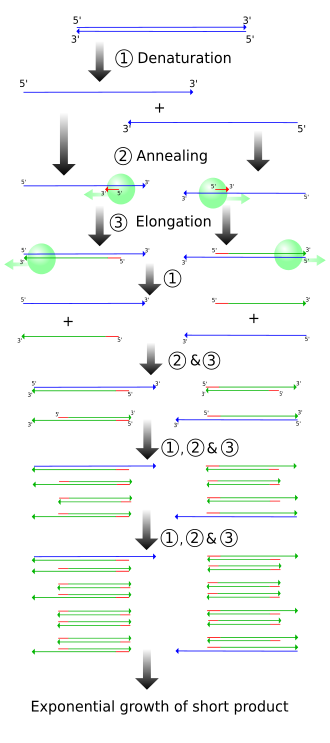
Diagram of the PCR cycle, showing denaturation (~95 °C), annealing (~55–65 °C), and extension (~72 °C) with primers and a thermostable polymerase. The figure also indicates how the target fragment is duplicated each cycle, driving exponential amplification. Temperatures shown may vary with primer Tm and enzyme used (extra procedural detail beyond the core syllabus). Source.
Thermocycler: A programmable machine that precisely controls temperature changes to enable the cyclic steps of PCR.

Photograph of a quantitative PCR thermocycler (Stratagene Mx3000). Thermocyclers automate the precise temperature cycling needed for denaturation, annealing, and extension. Display panels and 96-well blocks vary by model (instrumental details exceed the OCR core but aid recognition). Source.
Variations of PCR
Refinements and Adaptations
Several modifications of the basic PCR technique have been developed to suit different analytical or diagnostic purposes:
RT-PCR (Reverse Transcriptase PCR) – converts RNA into complementary DNA (cDNA) before amplification, useful for studying gene expression and detecting RNA viruses.
qPCR (Quantitative or Real-Time PCR) – monitors DNA amplification in real time using fluorescent markers, enabling quantification of the initial DNA amount.
Multiplex PCR – uses multiple primer pairs to amplify several DNA targets simultaneously, saving time and resources.
Nested PCR – improves specificity by using two sets of primers in successive reactions to reduce non-specific amplification.
Applications of PCR
1. Forensic Analysis
PCR is crucial in forensic science for amplifying DNA from trace biological evidence such as hair, saliva, or blood. The amplified DNA can then be analysed using DNA profiling to identify individuals with high accuracy.
Enables the analysis of minute or degraded samples.
Assists in paternity testing, criminal identification, and disaster victim identification.
2. Medical Diagnostics
PCR allows detection of pathogens, genetic disorders, and mutations with exceptional sensitivity.
Infectious diseases: Detects viral or bacterial DNA/RNA (e.g., HIV, COVID-19, tuberculosis).
Genetic conditions: Identifies mutations linked to diseases such as cystic fibrosis or Huntington’s disease.
Cancer diagnostics: Detects oncogene mutations and aids in personalised medicine approaches.
3. Research and Biotechnology
PCR is foundational in molecular biology research, enabling:
Cloning of genes into vectors for recombinant DNA studies.
Sequencing of amplified DNA to identify genetic variation.
Gene expression analysis when combined with RT-PCR.
Environmental monitoring, detecting microbial DNA in soil or water samples.
Advantages and Limitations
Benefits of PCR
High sensitivity – can amplify DNA from extremely small samples.
Speed and efficiency – results can be obtained within hours.
Specificity – primers ensure only the target sequence is amplified.
Automation – modern thermocyclers and reagents simplify setup and replication.
Limitations
Contamination risk: Even trace DNA contaminants can lead to false results.
Quantitative limits: Traditional PCR is not inherently quantitative without qPCR.
Sequence knowledge: Requires prior knowledge of the target sequence to design primers.
Inhibitors: Substances in the sample (e.g., haemoglobin, humic acids) can hinder enzyme activity.
Molecular Basis of Amplification
Exponential Nature of PCR
Each cycle theoretically doubles the DNA quantity. After n cycles, the number of copies equals:
EQUATION
—-----------------------------------------------------------------
PCR Amplification (N) = N₀ × 2ⁿ
N = Number of DNA copies after n cycles
N₀ = Initial number of DNA template molecules
n = Number of amplification cycles (no units)
—-----------------------------------------------------------------
This exponential replication underpins the power of PCR to detect and analyse even a single DNA molecule in complex biological samples.
Importance in Modern Biology
PCR has become indispensable in genomic research, forensic science, and clinical medicine. Its ability to amplify DNA with speed, precision, and sensitivity enables vital applications across diagnostics, genetic engineering, and evolutionary studies, fully addressing the OCR specification requirement to “explain the polymerase chain reaction and its application in amplifying DNA for analysis and diagnostics.”
FAQ
Taq polymerase is derived from the thermophilic bacterium Thermus aquaticus, which lives in hot springs. This enzyme remains active and stable at high temperatures used during PCR, particularly the denaturation stage at around 95°C.
DNA polymerases from most organisms would denature at these temperatures, halting the reaction. Using Taq polymerase ensures that DNA synthesis can proceed efficiently through multiple heating and cooling cycles without the enzyme needing to be replaced after each cycle.
Because PCR can amplify even trace amounts of DNA, contamination is a major concern. Scientists minimise this risk by:
Using sterile pipette tips and reagents.
Setting up reactions in laminar flow hoods or separate rooms.
Including negative controls (reactions without template DNA) to detect contamination.
Employing UV sterilisation and DNA-degrading enzymes to destroy stray DNA before preparation.
The annealing temperature depends primarily on primer length and base composition.
Short primers or those with more adenine-thymine (A–T) bases require a lower temperature because A–T pairs have fewer hydrogen bonds.
Longer primers or those rich in guanine-cytosine (G–C) pairs need a higher temperature for stable binding.
An incorrect annealing temperature can lead to non-specific binding (too low) or failure to bind (too high), reducing amplification efficiency.
Magnesium ions (Mg²⁺) act as cofactors for DNA polymerase, ensuring proper enzyme activity and stabilising primer-template binding.
Too little Mg²⁺ results in poor amplification because the enzyme cannot function effectively.
Too much Mg²⁺ can cause non-specific amplification or errors in base pairing.
Optimising magnesium concentration is therefore crucial for achieving high yield and specificity of the target DNA sequence.
PCR can amplify regions of DNA that contain known mutation sites. Once amplified, the DNA can be analysed using:
Restriction enzyme digestion – to identify changes that alter recognition sites.
Gel electrophoresis – to visualise differences in fragment size.
Sequencing – to determine the exact base change.
These methods allow detection of single-nucleotide mutations or deletions associated with genetic diseases, making PCR a powerful diagnostic and screening tool.
Practice Questions
Question 1 (2 marks)
State two essential components required for the polymerase chain reaction (PCR) and describe the function of each.
Mark Scheme:
1 mark for each correct component and its correct function.
Possible answers include:
Primers – short DNA sequences that bind to the target DNA to initiate replication. (1 mark)
DNA polymerase (Taq polymerase) – enzyme that synthesises new DNA strands by adding nucleotides to primers. (1 mark)
Nucleotides (dNTPs) – building blocks used by the enzyme to form new DNA strands. (1 mark, alternative to above if combined with another correct point)
Question 2 (5 marks)
Describe the main stages of the polymerase chain reaction (PCR) and explain how this process results in the amplification of a DNA fragment.
Mark Scheme:
Award marks for the following points:
Denaturation stage – DNA is heated to around 95°C to separate the double-stranded DNA into single strands by breaking hydrogen bonds. (1 mark)
Annealing stage – temperature is lowered to 55–65°C to allow primers to bind (anneal) to complementary sequences on the DNA template. (1 mark)
Extension stage – temperature is raised to around 72°C for Taq polymerase to add nucleotides, synthesising new complementary DNA strands. (1 mark)
Cycling – the three stages are repeated multiple times (typically 30–40 cycles), doubling the amount of DNA each cycle. (1 mark)
Exponential amplification – repeated cycles lead to millions of copies of the target sequence, enabling analysis from a minute starting sample. (1 mark)
Maximum 5 marks. Partial credit may be given for accurate descriptions of some stages without explicit temperature values.

