AP Syllabus focus:
‘Explain how the specific sequence of amino acids in a polypeptide defines its primary structure and influences the protein’s overall three-dimensional shape.’
Proteins begin as linear chains of amino acids. In AP Biology, the key idea is that the exact order of amino acids is information: it determines what the protein can fold into and, therefore, what it can do.
Primary Structure: The Sequence as Biological Information
Primary structure refers to the unique amino acid sequence of a polypeptide. Because different amino acids have different chemical properties, the sequence determines which parts of the chain can interact during folding, shaping the protein’s final form.
Primary structure: The unique, linear sequence of amino acids in a polypeptide (from one end of the chain to the other), held together by covalent peptide bonds.
Primary structure is often described as the “code” for a protein’s shape because the order of amino acids controls where particular chemical features (from side chains) occur along the chain.
What Counts as the “Sequence” in a Polypeptide?
To avoid ambiguity, biologists describe sequence in a consistent direction along the polypeptide:
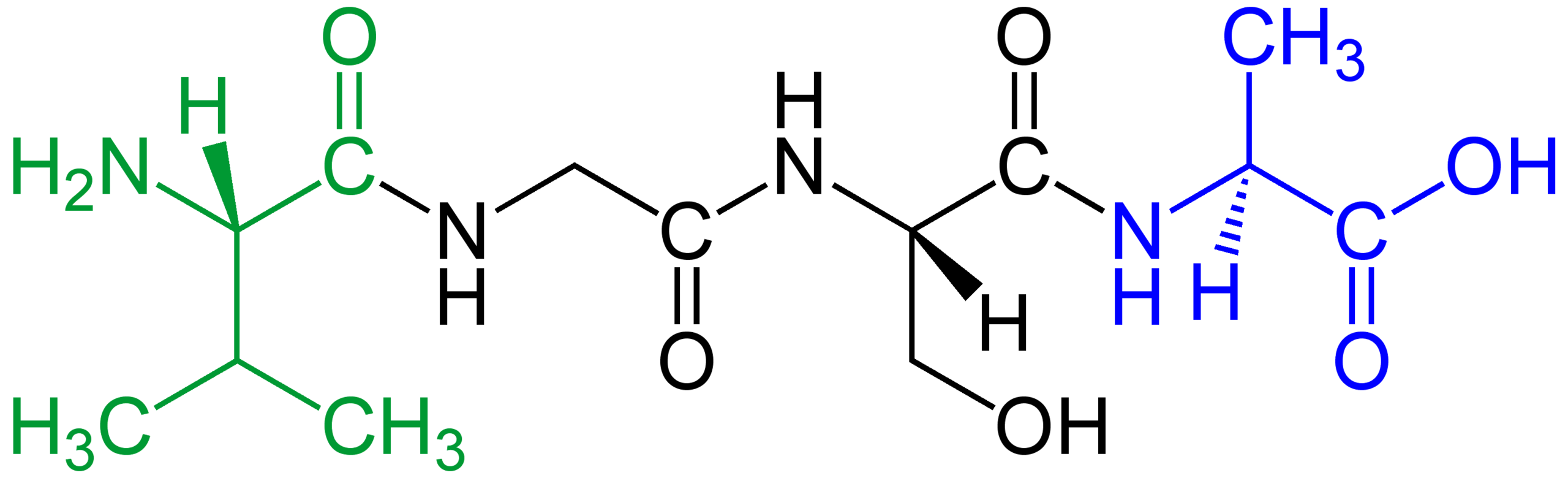
A tetrapeptide shown as a structural formula with the N-terminus highlighted in green and the C-terminus highlighted in blue. This emphasizes that “sequence” is not just the set of amino acids present, but their order from the amino end to the carboxyl end. Source
Amino (N) terminus and carboxyl (C) terminus: The two ends of a polypeptide; the N-terminus has a free amino group, and the C-terminus has a free carboxyl group.
Between these ends, each amino acid contributes to the backbone plus a distinctive side chain (R group). Even a single change in the sequence can shift where key side chains occur, which can alter folding outcomes.
How Sequence Influences Overall 3D Shape
The specification emphasizes that the specific sequence defines primary structure and influences the protein’s overall three-dimensional shape. This influence happens because:
The sequence determines the distribution of chemical properties along the chain (for example, which positions carry bulky, small, polar, or charged side chains).
Those properties control where the chain can form stabilising interactions during folding (such as attractions or repulsions among side chains, or interactions with water).
The resulting fold produces functional regions (such as binding pockets or interaction surfaces) that depend on precise positioning of amino acids.
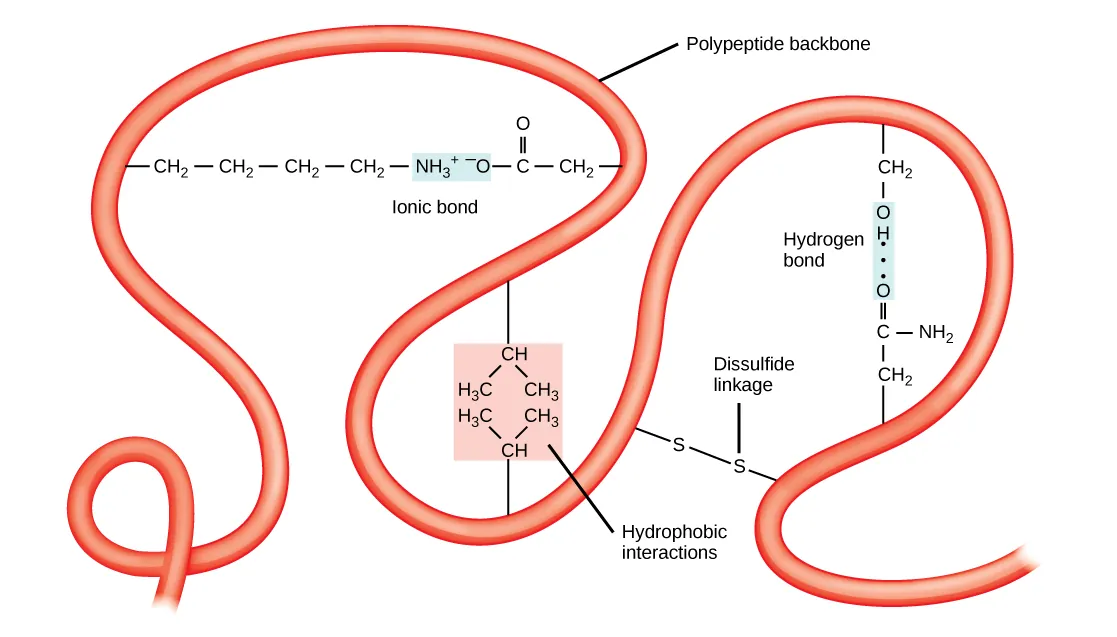
A folded polypeptide annotated with multiple side-chain interactions that stabilize tertiary structure, including hydrophobic interactions, ionic bonds, hydrogen bonds, and disulfide linkages. The figure makes clear that amino acid sequence matters because it determines which R groups are positioned to form these specific stabilizing interactions. Source
Primary structure does not directly describe the final shape, but it constrains and guides folding by setting the “starting conditions” for all downstream structure. In other words, different sequences tend to fold into different shapes because they present different side-chain patterns to the folding environment.
Why Small Sequence Changes can Matter
A polypeptide’s shape relies on many coordinated interactions. Sequence changes can therefore have effects such as:
No major change if the substitution preserves similar chemical properties and does not disrupt a critical region.
Local shape disruption if the change alters spacing, flexibility, or side-chain size in a region that must pack tightly.
Major loss or change of function if the change occurs at a position essential for activity (for example, a key binding or recognition site).
Levels of Sequence Specificity Within a Protein
Not every position in a protein is equally sensitive to change. Understanding this helps explain why primary structure is so informative.
Critical residues: amino acids at positions where a change strongly affects folding or function.
Tolerant regions: positions where multiple amino acids can substitute with minimal effect.
Sequence patterns: short, characteristic arrangements of amino acids that are associated with particular structural or functional features.
A common way to describe an amino acid within a chain is as a “residue,” meaning one amino acid unit after it has been incorporated into the polypeptide.
Primary Structure vs. Shape Changes Caused by Environment
Primary structure is maintained by covalent bonds along the backbone. Many conditions that alter protein shape (for example, changes in temperature, salt concentration, or pH) typically disrupt weaker interactions that stabilise folding rather than breaking the covalent backbone.
As a result:
############################
page_url: https://www.pearson.com/channels/biochemistry/learn/jason/protein-structure/anfinsen-experiment
image_identifier: Anfinsen Experiment schematic image (on page)
A schematic summary of the Anfinsen experiment showing that denaturation disrupts a protein’s native 3D conformation (and function), while removal of denaturants can allow refolding and recovery of activity. This provides evidence that primary structure contains the information needed for folding, even though environmental conditions can temporarily alter shape.
############################
A protein can lose its 3D shape while keeping the same primary structure.
Restoring conditions may sometimes allow the same primary structure to refold toward its original shape, because the sequence still encodes the folding tendencies.
FAQ
Common approaches include tandem mass spectrometry (MS/MS) to infer sequence from fragment masses.
Edman degradation can sequence short peptides, often used alongside enzymatic digestion to break proteins into manageable pieces.
Because the genetic code is degenerate: multiple codons can specify the same amino acid.
A DNA change may therefore not change the amino acid sequence, leaving primary structure unchanged.
A conservative substitution replaces an amino acid with one of similar chemical character, often causing less disruption.
A non-conservative substitution swaps to a very different side chain, more likely to alter folding tendencies.
Proteins from related species often have similar sequences due to shared ancestry.
Comparing sequence differences can help estimate relatedness, especially in highly conserved proteins.
Primary structure usually refers to the amino acid order specified by translation.
Some modifications (and proteolytic cleavage) can create a “mature” sequence that differs from the initial translated chain, affecting how the final protein behaves.
Practice Questions
Explain what is meant by the primary structure of a protein. (2 marks)
States that primary structure is the linear sequence/order of amino acids in a polypeptide (1)
Mentions amino acids are linked by peptide bonds / covalent bonds along the backbone (1)
A protein differs from a related protein by a single amino acid substitution. Explain how this change in primary structure could alter the protein’s overall three-dimensional shape and function. (5 marks)
Identifies that primary structure is the amino acid sequence and a substitution changes that sequence (1)
Explains that different amino acids have different chemical properties/side chains (1)
Links altered side-chain properties/positioning to changed interactions during folding (1)
Explains that changed folding can change the final 3D conformation (1)
Links altered 3D shape to altered function, e.g., changed binding site/interaction surface (1)

